
A targeted curation approach is necessary to transfer published data generated by primarily low-throughput techniques into interaction databases. Meaningfully representing this information remains far from trivial and different databases strive to provide users with detailed records capturing the experimental details behind each piece of interaction evidence. Network graphs generated from this data provide an understanding of the relationships between different proteins in the cell, and network analysis has become a widespread tool supporting -omics analysis. Molecular interaction databases are essential resources that enable access to a wealth of information on associations between proteins and other biomolecules. In this manuscript, we introduce these two algorithms and provide community access to the tool suite, describe examples of how these tools are useful to selectively present molecular interaction data and demonstrate a case where the algorithms were successfully used to identify a systematic error in an existing dataset. We have developed both a merging algorithm and a scoring system for molecular interactions based on the proteomics standard initiative-molecular interaction standards. Merging and scoring of data are commonly required operations after querying for the details of specific molecular interactions, to remove redundancy and assess the strength of accompanying experimental evidence. Experimental data is captured in multiple repositories, but there is no simple way to assess the evidence of an interaction occurring in a cellular environment. This method takes a SwingConstants integer value, indicating a compass direction (this enables us to add additional CytoPanels in the future, if we decide that's necessary.The evidence that two molecules interact in a living cell is often inferred from multiple different experiments. To obtain one, use the CytoscapeDesktop.getCytoPanel() method. If you are a core Cytoscape coder or a Plugin writer, here are a few tips to get started.Īs noted above, the Cytoscape Desktop contains three default CytoPanels.
The CytoPanel API is straightforward and fully documented. The end-user can show / hide any panel via the new CytoPanel menu, or via the keyboard accelerator short-cuts. By default, only the Control CytoPanel will appear, and it will automatically contain the network list and bird's eye view component.
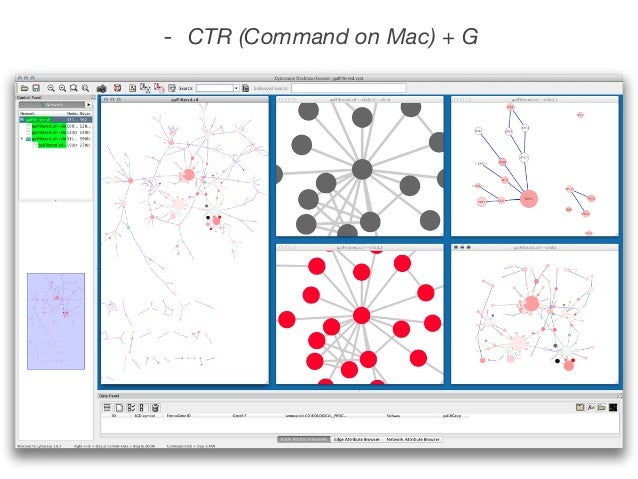
For example, in the screenshot below, the user has chosen to float it:Ĭytoscape 2.2 now includes three CytoPanels: CytoPanel 1 ('Control' appears on the left), CytoPanel 2 ('Data' appears on the bottom), and CytoPanel 3 ('Results' appears on the right). The user can then chose to resize, hide or float the left CytoPanel.

When you click on a node, the node details appear directly in the left CytoPanel. For example, the image below shows a screenshot of the latest BioPAX Plugin. Using the CytoPanel API, we can now show these node details in an embedded CytoPanel, and present a more integrated experience to the user. For example, in Cytoscape 2.1, the cPath Plugin enables users to click on a node and immediately view node details in a pop-up window.
#CYTOSCAPE TUTORIAL WINDOWS#
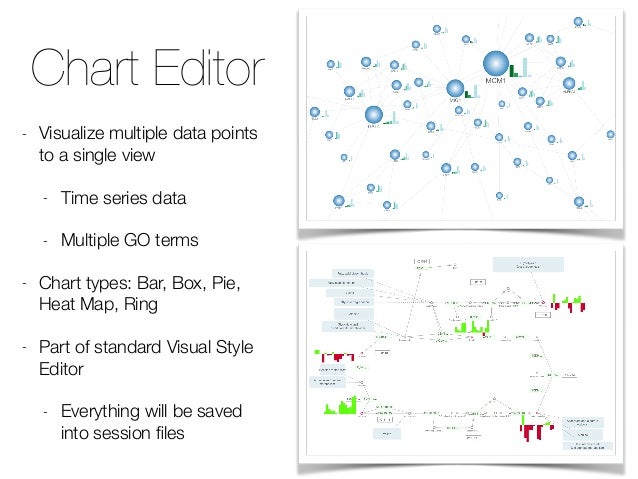
We built the CytoPanel API to cut down on the number of pop-up windows within Cytoscape, and create a more unified user experience. This feature is new in 2.2ĬytoPanels are floatable / dockable panels, which will be available in Cytoscape 2.2.
#CYTOSCAPE TUTORIAL HOW TO#
This page describes what CytoPanels are, how to use them, and how to programmatically access them.


 0 kommentar(er)
0 kommentar(er)
